MICROBIOMA - To be or not to be diseased: the effects of microbiome on parasitic infections in farmed seabream and sea bass
PTDC/MAR-BIO/0902/2014 – POCI-01-0145-FEDER-016550
Europeans are major consumers of seafood. However, only 20% of their requirements are met by European aquaculture production. In order to meet this demand, the sector will face a number of challenges. One such challenge will be dealing with infectious diseases which have major economic impacts.
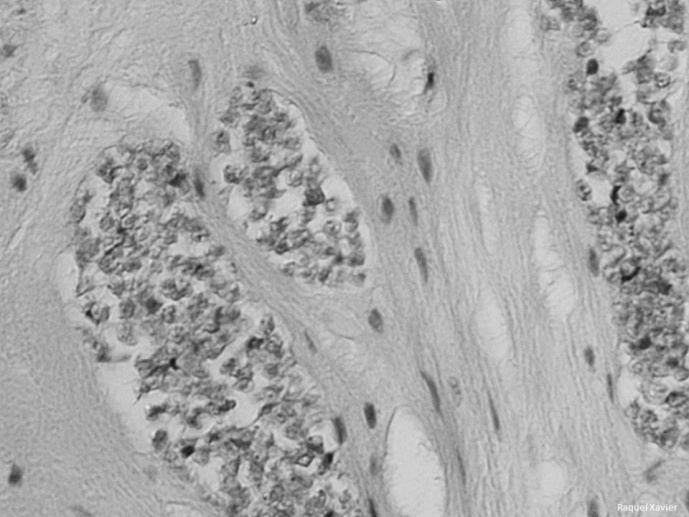
There is growing evidence that commensal microbes play a key role in immune response, and that differences in the microbiome can account for differential susceptibility to disease. In mammals huge research effort has been placed in deciphering microbiome-immune response interactions; however, in fish such research is still in its infancy. Still, pioneering work demonstrated that the stress caused by fish farming practices can induce changes in the skin microbiome of fish, with pathogenic bacteria becoming more predominant whereas beneficial bacteria, with an associated probiotic and/or antimicrobial function, decreased. Furthermore, changes in microbiome composition have been linked with susceptibility to disease as microbial commensals are known to prevent colonization by opportunistic pathogenic bacteria. For this reason, monitoring changes in microbiome has been suggested to predict disease outbreaks. Next Generation Sequencing (NGS) fills the technical gap to conduct such work, and the tools necessary to study entire microbial communities are now available. The main aims of this project are to characterise the commensal microbiota of farmed fish, and decipher their role in susceptibility to parasitic infections. In this project we will use state of the art NGS technology to perform a metagenomics analysis of microbial assemblages and transcriptomic analysis of host before and after infection with different pathogens. Specifically, we aim to unravel whether inter-individual variation in microbiome composition and function can be related to disease-susceptible, -tolerant and –resistant phenotypes; and also determine the role of host genetic background in microbiome composition.
Dates:
01 mai 2016 - 31 dez 2019
Total funding: 164 905,00 €
FEDER support: 140 169,25 €
National Funding: 24 735,75€
Joanne Cable, Ricardo Bruno de Araújo Severino